
Prof Dr Martin Simon / Molecular Cell Biology and Microbiology
Photo: Sebastian Jarych
A new diagnostic marker and the great mystery in biology
Molecular biologist and cell researcher Martin Simon on the significance of this year's Nobel Prizes for Medicine and Chemistry
Every year in October, the eyes of the world will be on Sweden when the Nobel Prize Committee honours international scientific work. However, the achievements of the prize-winning researchers are usually incomprehensible to the nonprofessional, as their focus is often too specialised. Wuppertal-based molecular biologist and cell researcher Martin Simon has been following this year's Nobel Prize awards in the fields of medicine and chemistry with great interest and explains the many opportunities that this opens up for research and future therapeutic approaches.
MicroRNAs
The 2024 Nobel Prize in Medicine will go to the US-Americans Victor Ambros and Gary Ruvkun for the discovery of microRNAs (small, non-coding RNA molecules) and their role in gene regulation. "I'll start with the genes," Simon begins, "because they are part of our genetic material and are switched on and off. This is how the different cell types in our body come about." Liver cells appear differently, for example, and have a different gene expression pattern than kidney, skin, epithelial or blood cells. "There are 23,000 genes in the human body, and different genes are always switched on and off, which allows us to form different organs and tissues. However, the gene must somehow be told whether it should be switched on or off, and this means that a protein is created from the DNA or not. This switching on and off is called gene regulation." MicroRNA is an essential component in this process. Although such small pieces of RNA, as Simon calls them, have been known for a long time, nothing was known about their gene regulatory abilities and they were categorised as a 'waste product'. However, when microRNA was analysed in more detail, it was eventually discovered that it has a negative impact on gene regulation. "If there is a matching microRNA for a gene, this gene is switched off and no protein is produced. It blocks protein synthesis."
To better categorise the significance of this discovery, Simon explains that it is like standing in front of a device and not knowing how it works. However, now we have found the 'switch' and can explain why things work this way or that. "The description of microRNAs has shown how all our cells do this and there are estimates that, in the most optimistic case, assume that around 60% of our genes are regulated by them. Now researchers can explain why, for example, this or that protein appears in the liver cell but not in the kidney cell, because they can see whether the microRNA prevents the formation of this protein. This is the switch that the two Nobel Prize winners have found and defined."
Research on the nematode provides insights for the human organism
The two award-winning scientists first discovered microRNA during their research on the nematode (C. elegans), which plays an important role in many research projects. "The nematode is a genetic model organism that has been known since the 1920s. It has enormous potential because it is an animal with many cells. It is possible to differentiate cell types, it has individual organs, but it is sufficiently primitive to be easily analysed and cultivated in large and rapid numbers of generations." A worm has just 800 cells and there are also methods for separating each individual cell of a worm and analysing it separately. "Then you have all organs and can group them. That would no longer be feasible with a mouse in terms of the effort involved. They are also used for toxicological analyses or the investigation of DNA damage. This worm really is an all-rounder for research and this has already been demonstrated at previous Nobel Prize awards." Back in 2006, a prize was awarded to two researchers who had worked with this worm. "So today is the second Nobel Prize in medicine to go to the first description of small RNAs for work with C. elegans. All these insights gained with this worm, and that is the special thing, can be transferred to the medical aspects of humans, and this is a particularly good example to illustrate the necessity for basic research."
MicroRNA can be used to better understand diseases
MicroRNAs are not to be confused with the mRNA made famous by Covid-19 vaccines - messenger RNA.. "With microRNA, we can now better understand diseases because gene expression is out of control in many diseases, especially cancer. We are beginning to understand how cancer develops in the first place and how this gene expression is misdirected, i.e. why are genes that are not switched on in normal liver cells now switched on in a liver carcinoma?" With this knowledge, it is then possible to understand how this cancer develops and why it is the way, it is. "We can see that many microRNAs are regulated differently in cancer. There are cancer-specific microRNAs present. The important thing is that these microRNAs are dysregulated here, because nature didn't make them to cause cancer." The scientists could now deduce from these dysregulations how quickly a specific therapy should be used or which kind of therapy would be appropriate. "This is very important for the therapy, because I may still have a longer time factor to treat the disease locally or have to do something very quickly to prevent metastasis. It has therefore also become a diagnostic marker." The research also opened up the possibility of using prognostic markers, i.e. acting in advance. "One example of such a prognostic marker is microRNA106b, because a genetic change in this RNA is associated with a higher risk of liver carcinoma. In microRNA, we have a prognostic marker, a diagnostic marker and we have an approach to intervene at these sites. Thus, we gained a therapeutic tool. We can block microRNAs or add them to cells that no longer produce them. We can intervene in this RNA system from the outside and influence the regulatory function, i.e. "we can flick the switch ourselves". Furthermore, since 2017, there have already been nine authorisations in the EU for active substances based on regulatory RNA."

Nobel Prize medal, public domain
A new world of protein structures
In the same week that the Nobel Prize for Medicine was awarded, the committee also announced the prize for chemistry to the scientists David Baker (USA) and Demis Hassabis and John Jumper (Great Britain). Back in 2003, Baker developed a computer programme that was able to design new proteins, thus opening up a new world of protein structures. "Proteins in themselves are the essential component of life, i.e. they are one of the main components of cells. They provide a scaffold structure and offer enzymatic activity to metabolise substances. All proteins are made up of different combinations of 20 building blocks, the amino acids, which are linked together in a linear chain. Now there are amino acids that are large, others are small, some are positively charged, and others are negatively charged." Logically, the composition of these amino acids is decisive for the properties of the protein as a whole. However, if you have a positive and a negative amino acid, the protein does not remain linear, but folds together. This creates a three-dimensional structure that has long been a mystery in biology. "If you imagine that there are proteins that are made up of several thousand amino acids, there could theoretically be so many different interactions between these individual building blocks. Nevertheless, you have to know the structure in order to understand the function of the protein. This used to be a huge effort. You had to crystallise the proteins, i.e. evaporate them and develop a crystal structure. And then you could use X-ray structure analysis or NMR analysis to find out how this individual protein is folded, i.e. you can create a 3D model." Even with this complex procedure, it was not always possible to crystallise all the proteins, even after several years of work. In this context, it is interesting to note that the Nobel Prize Committee has already awarded several prizes for research into the chemical analysis of protein structures in the past. With this year's award, however, a completely different approach is now coming into play. "An AI-based tool is used to calculate these interactions," explains Simon, "The tool is called 'AlphaFold', which is AI-based software that predicts the structure of proteins based on the amino acid sequence, i.e. the linear sequence. This tool is also self-correcting, so it checks the result itself again and gradually improves its prediction." This software trains and checks with already known structures. The fantastic thing about this software is that it has been published as open source, i.e. it can be used free of charge for both research and business. "We can already work with it. We can enter our amino acid sequences and get the structure after a little computing time. There are already follow-up programmes. For example, 'AlphaFold 3' can also calculate whether two different proteins interact and bind to each other, so you can already go into the next dimension. This is a huge step forward. We have installed it and are now using it permanently." The programme works as follows: the user enters a sequence of letters of their amino acid sequences and after a calculation time, the computer displays the three-dimensional pictorial structure, which can then be rotated in all directions. "In the past, that would have been the work of an entire doctorate," laughs the researcher. This speeds up research many times over and it is now the case that these structures are already stored in the genome databases for all genes.
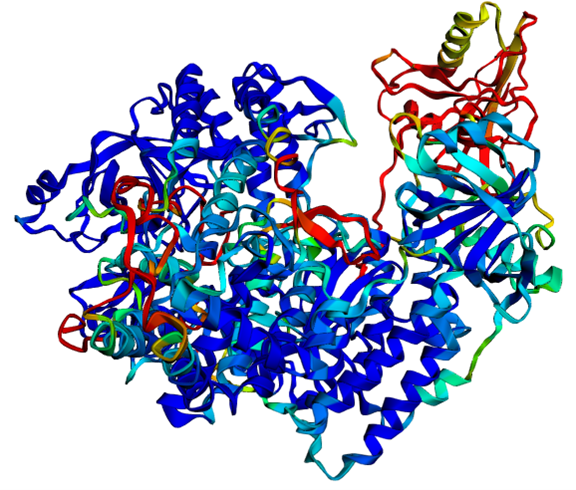
3D view of the AlphaFold2 calculated protein structure of an RNA-dependent RNA polymerase (P. tetraurelia RDR2)
AI also shows where it is unsafe
The scientists Hassabis and Jumper have created a gigantic database. This database, among other things, could also help to clarify antibiotic resistance or the use of enzymes to break down plastics.
"You can now have substances built with the software in a kind of design work," says the expert. As is so often the case, the user does not fully understand what the AI is doing. "With 'AlphaFold', for example, the areas in which the algorithm is unsure are coloured in. Blue means it is sure and red means it's not so sure. This is a key factor that must also be mentioned as a quality criterion. The programme leaves a mark in case of uncertainty."
Science creates impetus for the future
Nobel Prizes always mean honouring scientists and their research. However, what do these discoveries mean for everyday work?
"We are talking about the transfer of basic research, i.e. the Nobel Prize in Medicine has already been awarded twice to C. elegans researchers whose findings are transferable to humans." The Nobel Prize honours create new impetus for science with these awards. "This honour brings a transfer of new aspects into medicine," concludes Simon, emphasising: "We need new approaches. Just like with antibiotics. Although we have a further development of existing antibiotics, which are constantly being modified to avoid resistance, we are in an almost dead zone with classic antibiotics. We can only modify them, but we keep running into the next antibiotic resistance. Then we need new impetus in these fields. In addition, that could be e.g. viruses from basic research that infect bacteria. If we then modify them, we can break new ground again. With the Nobel Prize in Chemistry, though, it is the aspect that this new software 'AlphaFold' has already changed our daily lives in basic research because we can obtain solid results faster and more cost-effectively."
Uwe Blass
Prof Dr Martin Simon heads the Molecular Cell Biology and Microbiology group in the School of Mathematics and Natural Sciences at the University of Wuppertal.
